Genetic basis of quantitative traits
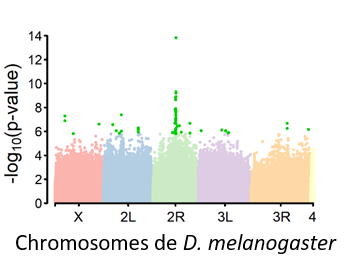
In recent years, I have learned to characterize the genetic basis of phenotype variation using genetic mapping (GWAS). This allowed me to participate in several projects using the genetic panel of Drosophila melanogaster DGRP to make GWAS. The GWAS with the DGRP is regularly undermined. However, unlike many models, the tractability of Drosophila allows to confirm the candidate genes provided by GWAS, making it a prime system to study the genetic basis of quantitative traits.
We determined the genetic basis of D. melanogaster’s resistance to two commonly used insecticides. Duneau et al. 2019 In collaboration with a team of toxicologists, one from population genetics and one from functional biology, this interdisciplinary study confirmed that the resistance genes, we validated using functional genetics, show selection signatures in nature on non target species. We report a potential role of Wolbachia in the efficacy of resistance genes.
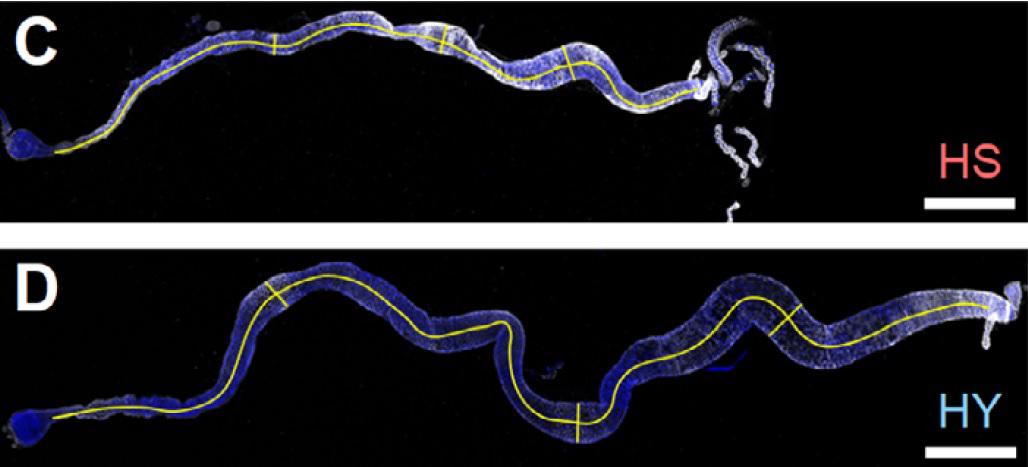
In another collaboration with Nicolas Buchon (Cornell University, USA) I studied the genetic basis of the variation in the plasticity of the morphology of the gut in response to diet. Bonfini et al. 2021
- In collaboration with Patricia Beldade and Elvira Lafuente we studied the genetic basis of the variation of plasticity in size and pigmentation in response to temperature (Lafuente et al., 2018, Lafuente et al., in prep). We have shown, among other things, that it is highly unlikely that there is a genetic basis for general phenotypic plasticity. Rather, we show that each phenotype responds plastically to a given element of the environment independently of the other phenotypes. This independence is made possible by a genetic basis specific to each phenotype, which therefore allows plasticity to evolve independently for each trait.